
Marine hypoxia and pelagic redoxclines
Oxygen-deficient (hypoxic) conditions within a water column can be found in a variety of marine systems. Typically they occur in coastal and marginal seas with permanent or temporal stratification or in the oxygen minimum zones near upwelling areas. Most prominent pelagic systems with anoxic conditions are the Black Sea, the Baltic Sea deeps, and the Cariaco Basin. In the world’s two largest brackish systems, the Black Sea and the Baltic Sea, stratification is due to freshwater input and the establishment of a halocline, which prevents mixing of upper oxygenated and deeper anoxic and sulfidic waters, leading to an intermediate layer with strong vertical redox gradients: the redoxcline. The redox gradient describes the vertical segregation of major electron acceptors and reduced products. It separates the upper oxygenated from anoxic bottom water and is, for this reason, an area of elementary biogeochemical transformations. In aquatic environments it is commonly referred to as the `pelagic redoxcline´ (to distinguish from sedimentary redoxclines) whereas the depth of the oxic-anoxic interface or of the first appearance of sulfide is usually defined as `chemocline´.
While anoxic conditions prevail in the Black Sea since about 8,000 years, the Baltic Sea is characterized by regularly occurring inflow events from the North Sea partly causing complete oxygenation of the anoxic deeps. In contrast, the CaribbeanCariacoBasin, which is a deep, isolated depression on the continental margin of Venezuela, is the largest, truly marine, anoxic basin, and permanent stratification is due to a strong temperature gradient. Despite these differences between the three systems, many similarities in biogeochemical processes and microbially mediated transformations within the carbon, nitrogen, and sulfur cycle have been revealed in recent years. Anoxic and sulfidic bottom water can also be found in many fjords with a sill as demonstrated by several studies describing the situation in the Framvaren Fjord (Norway) or NitinatLake (British Columbia). Similar mechanisms may also occur in lakes, where temperature or dissolved substances contribute to density stratification, often accompanied by hypoxic or anoxic deep-water conditions. However, major differences to marine systems exist for many physical, geochemical, and biological features in stratified lakes.
Related to hypoxia in marine systems are oxygen minimum zones (OMZs), which cover about 8 % of the ocean. They are different to the above mentioned anoxic systems as oxygen is usually not completely exhausted. However, in the core of OMZs the appearance of sulfide has also been observed temporally. Similarities of OMZs with the above mentioned anoxic marine areas can also be found with respect to several biogeochemical processes and the involved microbial key organisms.
Detailed knowledge about the present situation is also the key for understanding the past. For instance, the distribution of anoxic marine water columns was much wider during the Phanerozoic, the most extremes being developed during so-called Oceanic Anoxic Events, now witnessed in the deposition of black shales. In this context, investigating modern euxinic systems is of particular scientific interest to understand the development, extent and consequences of anoxic systems. Euxinic conditions have been suggested to be associated with times of global environmental changes associated with sometimes dramatic life extinctions, and hydrogen sulfide, that may have even reached the ocean surfaces in the past, is suspected to be involved in massively killing terrestrial and marine life. Therefore, understanding the development and processes involved in pelagic anoxia is of fundamental interest also for understanding extreme changes in paleo-ecological conditions.
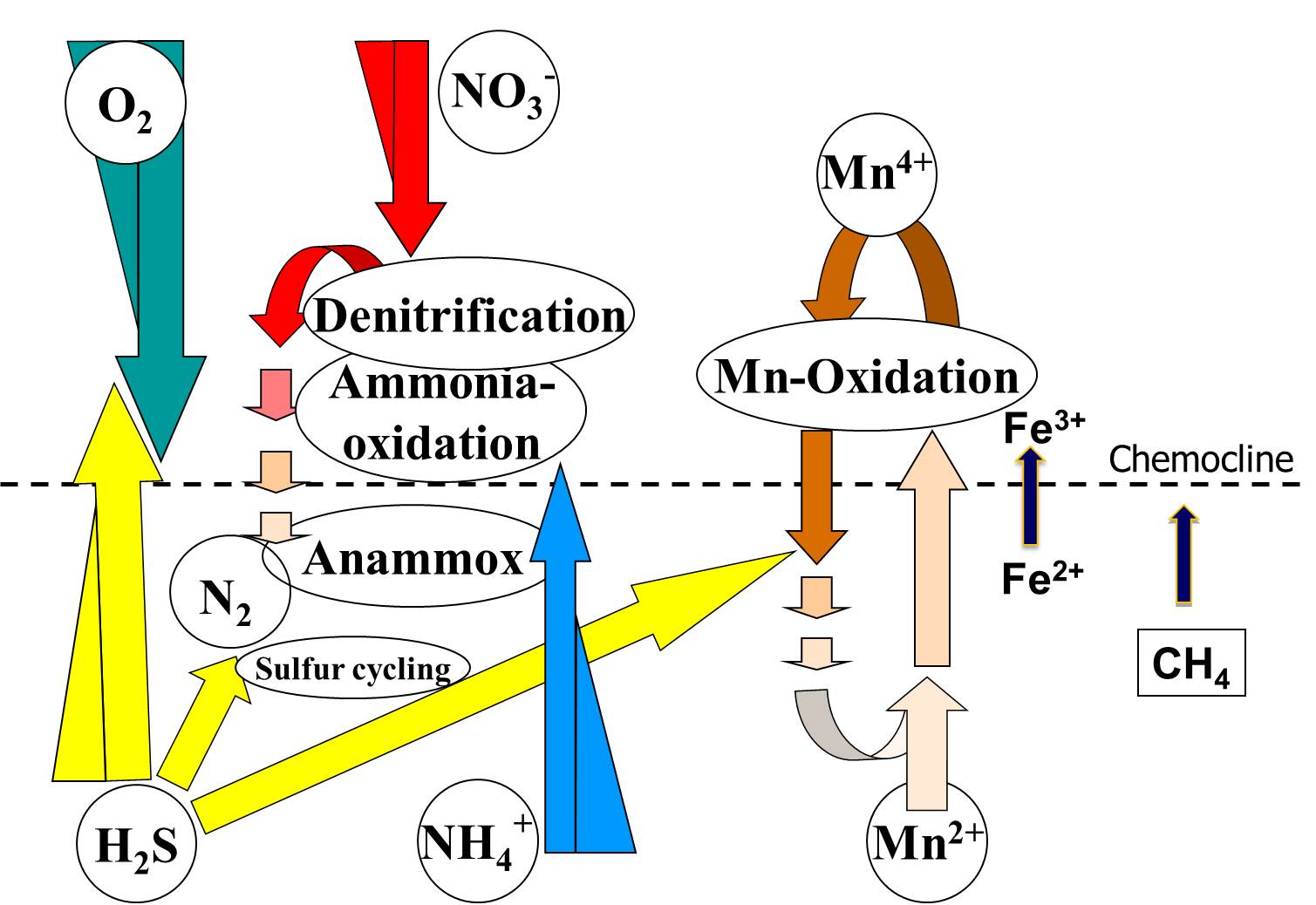
Figure 1. Some known major biogeochemical processes at marine or brackish pelagic redoxclines.
Within the marine science community there has been increasing interest in hypoxic systems in recent years because of their assumed global increase and their impact on biogeochemical cycles. For the microbiological community the study of hypoxic systems has been particularly rewarding because of the discovery of novel metabolic pathways and novel phylogenetic groups of microorganisms. The redoxcline of the central Baltic Sea is ideally suited as a model system to study principal mechanisms, biogeochemical processes and responsible organisms because (1) it shows many similarities to other marine anoxic systems with respect to hydrographic forces, biogeochemical processes and the structure of the microbial communities, and (2) the necessary infrastructure, logistics and profound knowledge is available within the different departments of the IOW. Interdisciplinary research will be carried out also by the Project Group “Pelagic Redoxcline” of the IOW, in which scientists from all departments participate for a better understanding of redoxcline processes.
Some highlights of our research
With the application of innovative molecular techniques many new insights into the composition and function of microbial communities in hypoxic systems and pelagic redoxclines have been revealed. Strong shifts in microbial composition occur particularly across the oxic-anoxic interface and even in sulfidic waters an astonishing diversity of prokaryotes and microbial eukaryotes (protozoa) has been discovered, nearly totally consisting of uncultivated taxa.
We could make significant progress in recent years in identifying functional groups of prokaryotes that could be considered as key players for distinct transformation processes in pelagic redoxclines of the Baltic Sea. However, these key players have also been found in hypoxic marine systems distributed around the globe, such as the Black Sea, the Cariaco Basin, or different oceanic OMZ.
Chemolithoautotrophic prokaryotes as key players for the nitrogen and sulfur cycles in Baltic and Black Sea pelagic redoxclines
Characteristic for pelagic redoxclines outside photic zones is the considerably high chemoautotrophic production (measured as dark CO2 fixation). On a yearly basis it can be in a similar range as surface primary production. The microbial community is dominated by chemoautotrophs, and their production supports an active redoxcline microbial food web, consisting of different groups of protists (flagellates, ciliates). Processes potentially supporting dark carbon dioxide fixation include aerobic and anaerobic ammonium oxidation, chemoautotrophic denitrification, aerobic methane oxidation, metal oxidation, hydrogen oxidation, disproportionation of sulfur compounds, and aerobic or anaerobic microbial sulfur oxidation.
An estimation of the abundance of chemolithoautotrophic prokaryotes was possible by combining dark CO2 fixation measurements with flow cytometric cell sorting (Fig. 2). Flow cytometric analyses of DNA-stained bacterioplankton revealed the existence of at least five different prokaryotic clusters, distinguished by side-scatter and DNA fluorescence, in water samples collected beneath the chemocline. Dark CO2 fixation in these clusters was determined by flow cytometric sorting after anaerobic incubations with NaH14CO3 tracer. Two clusters, representing about 30 % of total prokaryotes, were responsible for 65 to 100 % of the total dark fixation (Fig. 2). Calculated cell-specific carbon fixation rates for these clusters were in the range of 3.5 to 24.7 fg C cell-1 d-1 which suggested that these clusters were strongly dominated by chemolithoautotrophic prokaryotes. Mean cell-specific fixation rates reached > 10 fg C cell-1 d-1 in most cases, indicating relatively high growth rates (estimated doubling time 1-2 d) of chemolithoautotrophic prokaryotes. These results provided for the first time evidence for such high cell-specific CO2 uptake and considerable abundance of chemolithoautotrophic prokaryotes in a pelagic marine environment.
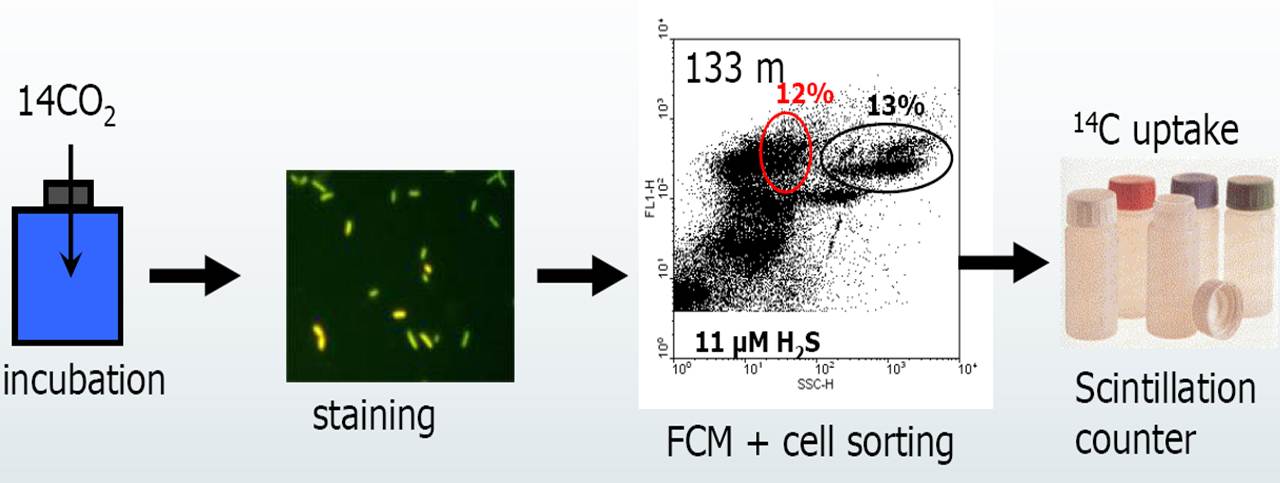
Figure 2. Principle of the flow cytometric approach. Prokaryotic groups were flow sorted for determining cell dark CO2 fixation rates. The flow sorted groups are: Bpl, prokaryotes with low nucleic acid content (LNA), LNA cells with high side scatter (LNA-hs), prokaryotes with high nucleic acid content (HNA) and high side scatter (HNA-hs), medium side scatter (HNA-ms), and low side scatter (HNA-ls). Arrows indicate the clusters of sorted cells. Dark CO2 fixing clusters are marked in red. Modified from Jost et al. (2008).
The assumption that epsilonproteobacteria are the main candidates for chemoautotrophy in pelagic redoxclines was proven by a whole set of experimental and molecular approaches. Results from 16S rDNA cloning revealed that 21% of the derived clone sequences (grouped together under the name “GD17”), had > 99% similarity to a previously described uncultured epsilonproteobacterium, closely related to Sulfurimonas denitrificans (Fig. 3).
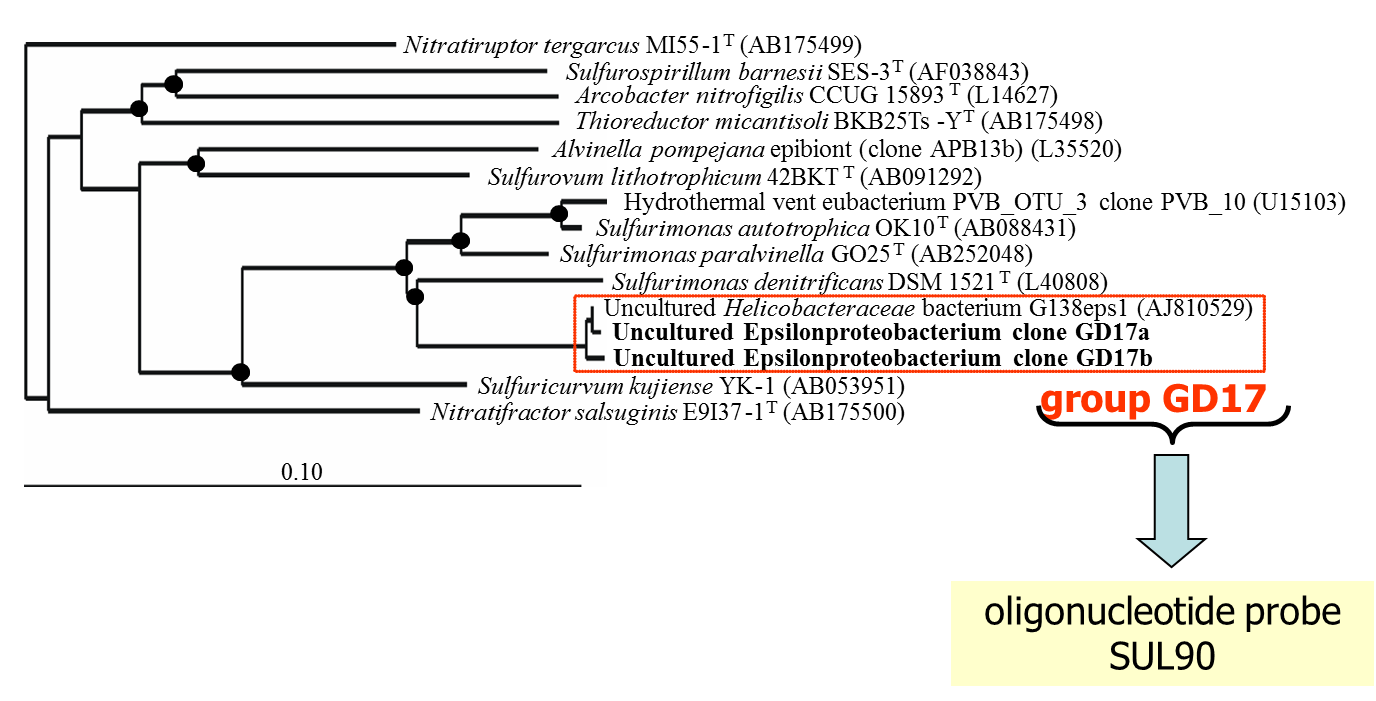
Figure 3. Unrooted tree showing relationships of subgroup GD17 and its closest phylogenetic relatives within the Epsilonproteobacteria. The framed area denotes subgroup GD17 originating from the central Baltic Sea as the target sequence of gene probe SUL90 (modified from Grote et al., 2007).
A specific gene probe against GD17 was developed and used for enumeration by CARD-FISH. GD17 cells were always detectable from the lower oxic to the sulfidic area, with maximal numbers around the chemocline, representing up to 20% of the total bacteria. Epsilonproteobacteria consisted nearly entirely of cells belonging to the distinct GD17 group. This suggested that GD17 was the best adapted epsilonproteobacterium within this ecological niche. We found high cell numbers of Epsilonproteobacteria also in pelagic redoxclines of the Black Sea, confirming their general importance in this habitat.
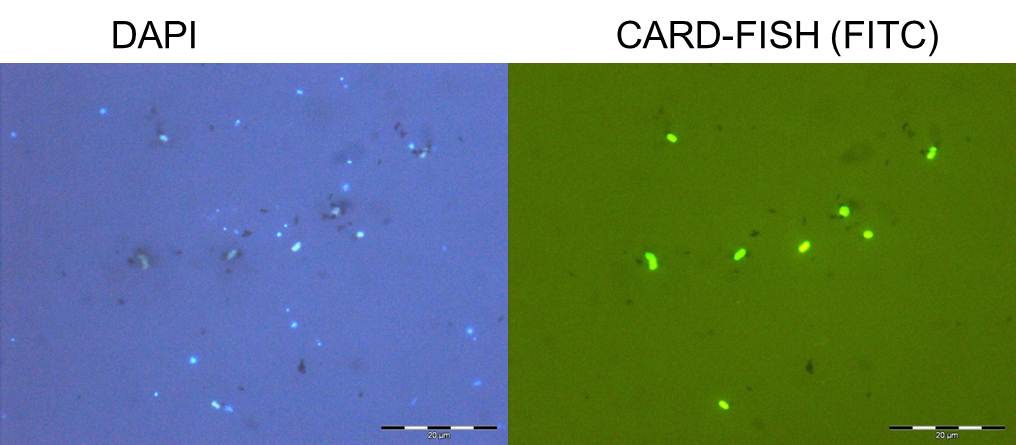
Figure 4. Whole cell hybridization (CARD-FISH) with probes for Epsilonproteobacteria and Sulfurimonas GD17, combined with microautoradiography of central Baltic Sea samples incubated with 14C-bicarbonate (see Grote et al. 2008).
In order to identify the major chemoautotrophic prokaryotes and also to prove that Epsilonproteobacteria are involved in CO2 fixation in the redoxcline, we combined [14C] bicarbonate incorporation studies with CARD-FISH analysis and microautoradiography (MARFISH). In absolute numbers, maximal 3 x 105 14CO2 fixing prokaryotic cells mL-1 were detected for a central Baltic Sea and 9 x 104 14CO2 fixing cells mL-1 for a Black Sea redoxcline. These numbers corresponded to 29 % and 12 % of total prokaryotic cell abundance, respectively. 14CO2 incorporating cells belonged exclusively to the phylum Bacteria. Among these, approximately 70 % in the central Baltic and nearly 100 % in the Black Sea (Fig. 5) were members of the Epsilonproteobacteria. Moreover, for the Baltic Sea the particular Sulfurimonas subgroup GD17 could be identified as the absolutely dominating chemoautotrophic organism.
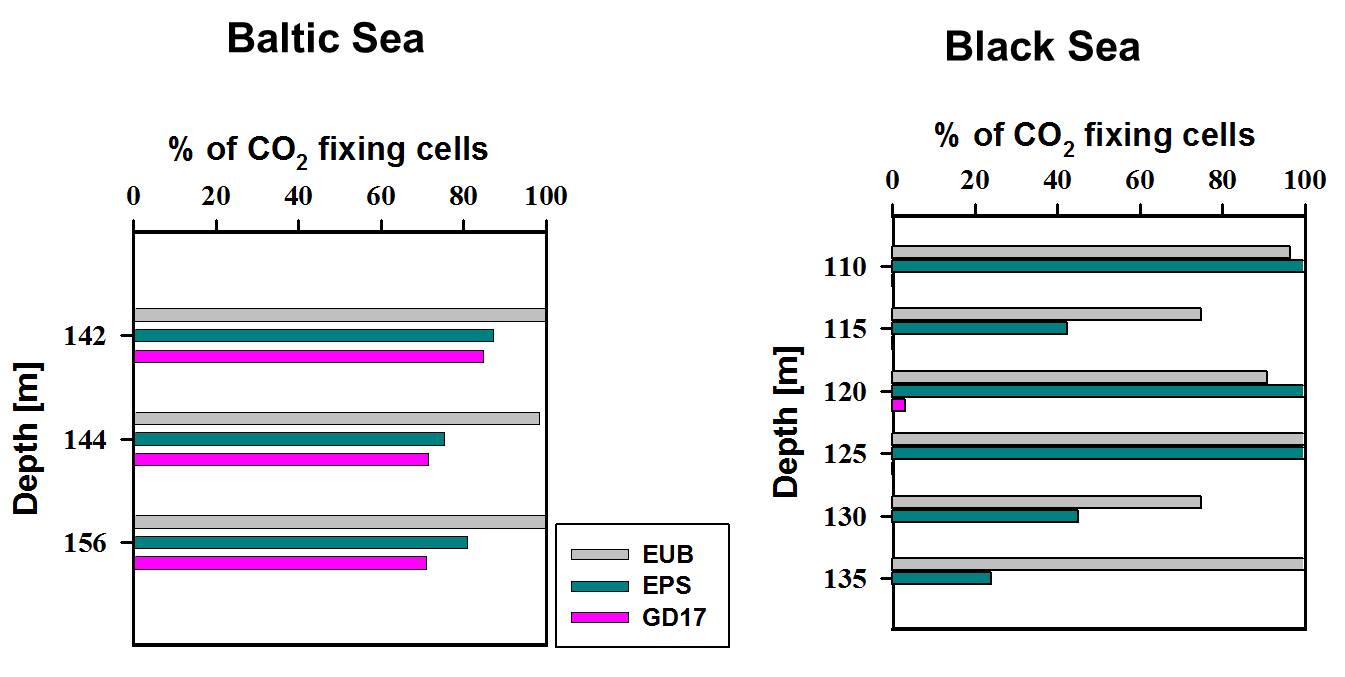
Figure 5. Relative proportion of 14CO2-assimilating prokaryotes belonging to Eubacteria (EUB), Epsilonproteobacteria (EPS), and group GD17, as determined by MARFISH for redoxcline samples from the Baltic and the Black Sea. Modified from Grote et al. (2008.)
Another promising approach to identify active chemoautotrophic prokaryotes is to use 13C-bicarbonat incubations with the stable isotope analysis of extracted RNA (SIP-RNA). Incubations for the RNA-SIP approach revealed that two different Gammaproteobacteria and three closely related Epsilonproteobacteria, again of the Sulfurimonas cluster, were actively dark CO2 fixing microorganisms (Fig. 6). After 72 h of incubation the 13C-label had been transferred to a potential bacterivorous ciliate related to Euplotes sp. Thus, RNA-SIP provided direct evidence for the contribution of chemolithoautotrophic production to the microbial food web in this marine pelagic redoxcline, emphasizing the importance of dark CO2 fixing Proteobacteria within this habitat.
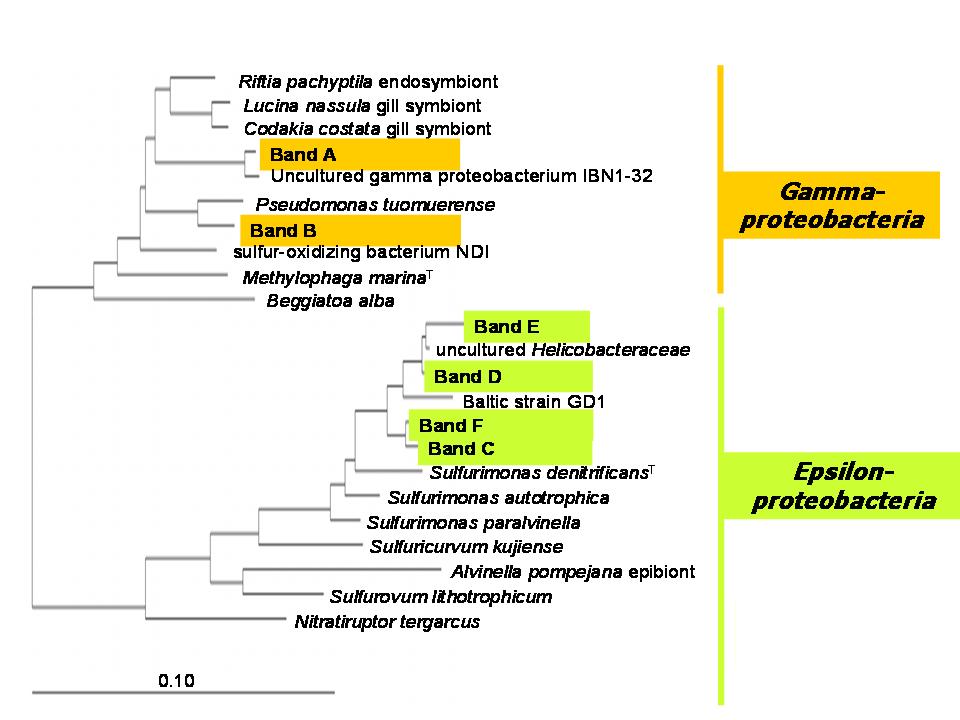
Figure 6. Unrooted phylogenetic tree of bacterial 16S rRNA sequences showing the affiliation of the identified bacterial SSCP bands, enriched in 13C, and their closest relatives within the Epsilon- and Gamma-proteobacteria. The tree was reconstructed using the neighbor-joining method based on com-parison of approximately 400 nucleotides. Only bootstrap values above 70% are shown. Bar, 10 substitutions per 100 nucleotides. Modified from Glaubitz et al. (2009).
Physiological and genomic studies of an isolated representative of the Baltic Sea Epsilonproteobacteria: Sulfurimonas gotlandica GD1
Model organisms from pelagic redoxclines have not been available until recently, which has contributed to a poor understanding of these microbes. We were able to isolate and culture one bacterium which proved to be a key player for chemoautotrophic denitrification in the redoxclines of the Baltic Sea: ´Sulfurimonas gotlandica´ str. GD1, belonging to the Epsilonproteobactera. Whole genome analysis and laboratory experiments revealed a high metabolic flexibility, suggesting a considerable capacity for adaptation to variable redox conditions. ´Sulfurimonas gotlandica´ str. GD1 was shown to grow chemolithoautotrophically by coupling denitrification with oxidation of reduced sulfur compounds and dark CO2 fixation. Metabolic versatility was further suggested by the use of a range of different electron donors and acceptors and organic carbon sources. The number of genes involved in signal transduction and metabolic pathways exceeds those of other Epsilonproteobacteria. Oxygen tolerance and environmental sensing systems combined with chemotactic responses enable this organism to thrive successfully in marine oxygen depletion zones. We propose that ´Sulfurimonas gotlandica´ str. GD1 will serve as a model organism in investigations that will lead to a better understanding how members of the Epsilonproteobacteria are able to cope with water column anoxia and the role these microorganisms play in the detoxification of sulfidic waters.
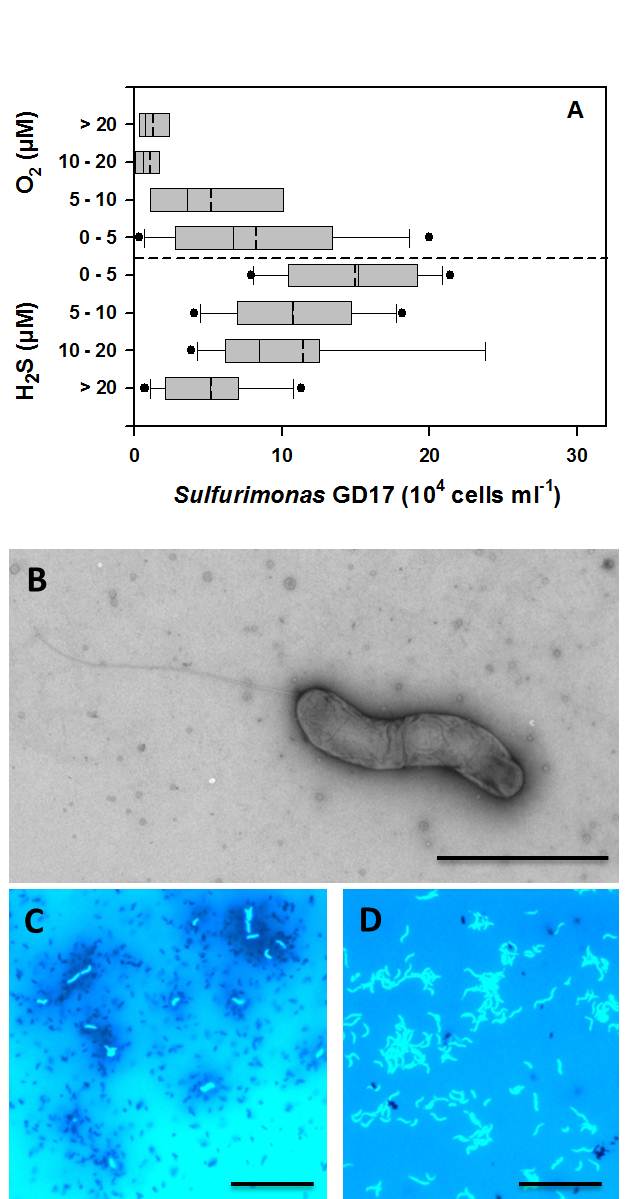
Figure 7: Sulfurimonas GD17 in Baltic Sea redoxclines (from Grote et al. 2012). A. Abundance of the Sulfurimonassubgroup GD17 as determined by CARD-FISH versus grouped concentrations of sulfide and oxygen in Baltic Sea redoxcline samples. Values for sulfide are expressed as negative oxygen concentrations. Box-and-whisker plots with outliers include median (solid) and mean (dashed) values within the boxes. Data from 89 samples collected from redoxclines in the central Baltic Sea during cruises between 2003 and 2009, including 35 data points from refs. (17, 20), is displayed. B. Electron micrograph of a phosphotungstic acid (PTA) negatively stained ´Sulfurimonas gotlandica´ str. GD1 cell cultivated on nitrate and thiosulfate demonstrating spirilla-like morphology and monopolar flagella. Bar - 2 µm. C. Fluorescence microscopy of DAPI-stained ´Sulfurimonas gotlandica´ str. GD1 cells grown on 14C-bicarbonate, followed by microautoradiographic incubation (17). Silver granules surrounding GD1 cells indicate 14C-bicarbonate, suggesting chemolithoautotrophic activity. D. Fluorescence microscopy of DAPI-stained ´Sulfurimonas gotlandica´ str. GD1 cells. Bar - 10 µm.
In vitro experiments and genome analysis revealed that ´Sulfurimonas gotlandica´ str. GD1 has a considerable metabolic versatility, capable of both chemoautotrophy (fueled by different electron donor and acceptor combinations) and utilization of organic substrates. Experiments confirmed the consumption of nitrate and hydrogen sulfide or thiosulfate and production of sulfate, suggesting that chemolithoautotrophic denitrification is an important respiratory pathway in this organism. ´Sulfurimonas gotlandica´ str. GD1 showed a pronounced ability to exploit a wide range of reduced sulfur compounds as electron donors, including elemental sulfur, sulfite, and hydrogen sulfide. Genome analysis revealed that the ´Sulfurimonas gotlandica´ str. GD1 chromosome contains a complete set of genes required for chemolithoautotrophy and encodes the components of the reductive citric acid cycle (rTCA) for inorganic carbon fixation, all reductases necessary to reduce nitrate to nitrogen gas, including NAP and NAR, and different enzyme systems for the oxidation of reduced sulfur compounds (Fig. 8). In addition to the sulfur reducing Sox multienzyme complex, several enzymes known to oxidize sulfite to sulfate via the intermediate adenosine-5´-phosphosulfate in a Sox-independent pathway were also encoded in the genome.
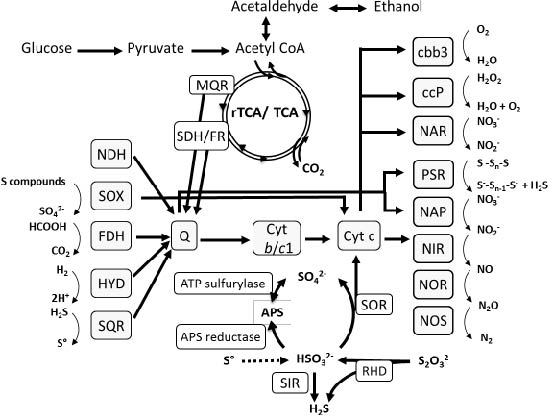
Figure 8: Theoretical model of the metabolic pathways and electron transport in ´Sulfurimonas gotlandica´ str. GD1.Enzymes catalyzing key reactions are indicated; reactions for which no enzyme was annotated in ´Sulfurimonas gotlandica´ str. GD1 but are predicted to occur are indicated by dotted lines.FDH: formate dehydrogenase, HYD: hydrogenase, SQR: sulfide quinone reductase, SOX: sulfur oxidation multienzyme complex, SOR: sulfite oxidoreductase, SIR: sulfite reductase, Q: quinone, Cyt b/c1: quinone cytochrome oxidoreductase, cbb3: cytochrome c oxidase, ccP: cytochrome c peroxidase, NAP/NAR: nitrate reductase, NIR: nitrite reductase, NOR: nitric oxide reductase, NOS: nitrous oxide reductase, PSR: polysulfide reductase, RHD: rhodanese-related sulfurtransferase, MQR: malate-quinone-reductase, SDH/FR: succinate dehydrogenase/ fumarate reductase. (Grote et al. 2012)
Diversity, phylogeny and ecological role of heterotrophic protists in the redox gradients of the central Baltic Sea
Our knowledge regarding the diversity and function of microorganisms in pelagic redoxclines is based mainly on data from prokaryotic communities. Comparable studies on microbial eukaryotes (protists) are still largely lacking. Our aim is to investigate the composition and functional role of phagotrophic protists (mainly flagellates and ciliates) along the redox gradients. To this aim, the quantitative species composition of phagotrophic protists is being examined by a combination of molecular techniques (18S DNA/RNA fingerprints, clone libraries, in situ hybridization, quantitative PCR), classical microscopical methods, and incubation experiments with natural assemblages and isolated organisms. In order to understand the impact of the oxygen gradients on the major mortality factors for planktonic prokaryotes, we also quantify mortality rates due to protist grazing and viral lysis along the gradients. The Baltic Sea redoxcline will serve as a model system to reveal a general understanding of bacteria-protist interactions at oxic-anoxic interfaces. Samples and experiments from other marine redoxclines will serve to test whether the patterns observed in the Baltic Sea can be generalized.
In situ investigations on redoxcline protist communities were are successfully complemented with studies on isolated organisms from the environment, which are used for ecophysiological investigations and species descriptions of morphological indistinguishable species. For example, we were able to isolate and cultivate several choanoflagellate strains belonging to lineages so far only known from environmental sequencing approaches. Quantitative experimental data for choanoflagellates from Gotland and Landsort Deep (Baltic Sea) indicate their preference for oxygen-depleted zones and thus their potentially high ecological relevance for Baltic Sea hypoxic waters. Cultivation of protists inhabiting hypoxic habitats has been unsuccessful in most cases, and has never yet succeeded for choanoflagellates, even though these small bacterivorous flagellates are known to be ecologically relevant in terms of biomass and abundance. Our preliminary ecological investigations have shown that one of the choanoflagellate strains is able to grow successfully under suboxic conditions in the laboratory and reach high cell abundances by very weak oxygen saturation.
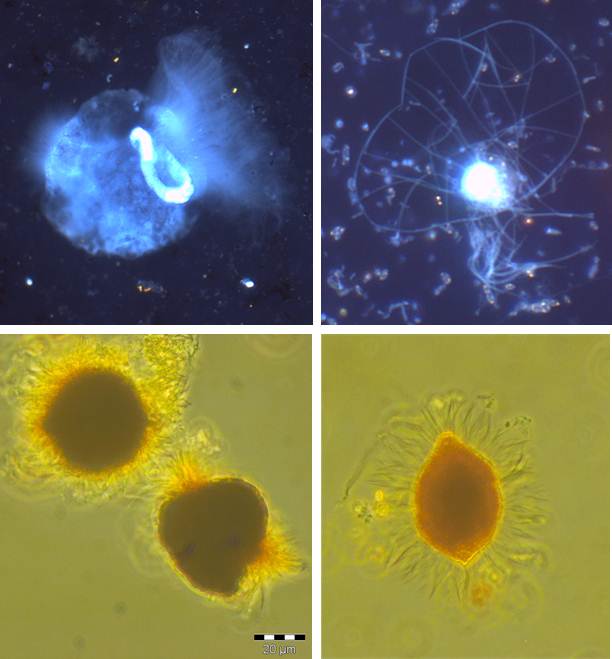
Figure 9: Microphotographs of protists from Baltic Sea redoxclines. Above: after DAPI staining; below: Lugol fixed samples (Photos by R. Anderson and C. Wylezich).
Publications resulting from our research on pelagic redoxclines
Anderson, R., Winter, C., Jürgens, K. (2012). Relevance of protist grazing and viral lysis as prokaryotic mortality factors for Baltic Sea oxic-anoxic interfaces. Mar. Ecol. Prog. Ser. (in press)
Bruckner, C.G., Mammitzsch, K., Jost, G., Wendt, J., Labrenz, M., Jürgens, K. (2012). Chemolithoautotrophic denitrification of epsilonproteobacteria in marine pelagic redox gradients. Environ. Microbiol. doi:10.1111/j.1462-2920.2012.02880.x
Grote, J., Schott, T., Bruckner, C.G., Glöckner, F.O., Jost, G., Teeling, H., Labrenz, M., Jürgens, K. (2012). Genome and physiology of a model Epsilonproteobacterium responsible for sulfide detoxification in marine oxygen depletion zones. Proc. Nat. Acad. Sci. USA 109: 506-510.
Feike, J., Jürgens, K., Hollibaugh, JT, Krüger, S., Jost, G., Labrenz, M. (2012). Measuring unbiased metatranscriptomics in pelagic aerobic ammonium oxidation zones of the central Baltic Sea based on a new in situ fixation system. ISME J. 6: 461-470.
Wylezich, C., Jürgens, K. (2011). Protist diversity in suboxic and sulfidic waters of the Black Sea. Environ. Microbiol. 13: 2939-2956.
Labrenz, M., Sintes, E., Toetzke, F., Zumsteg, A., Herndl, G. J., Seidler, M., Jürgens, K. (2010). Relevance of a crenarchaeotal subcluster related to Candidatus Nitrosopumilus maritimus to ammonia oxidation in the suboxic zone of the central Baltic Sea. ISME J 4, 1496-1508.
Glaubitz, S., Labrenz, M., Jost, G., & Jürgens, K. (2010). Diversity of active chemolithoautotrophic prokaryotes in the sulfidic zone of a Black Sea pelagic redoxcline as determined by rRNA-based stable isotope probing. FEMS Microbiol Ecol 74, 32–41.
Jost, G., Martens-Habbena, W., Pollehne, F., Schnetger, B., & Labrenz, M. (2010). Anaerobic sulfur oxidation in the absence of oxygen and nitrate dominates microbial chemoautotrophy beneath the pelagic chemocline of the Eastern Gotland Basin, Baltic Sea. FEMS Microbiol Ecol 71, 226-236
Glaubitz, S., Lueders, T., Abraham, W.-R., Jost, G., Jürgens, K. & Labrenz, M. (2009). 13C-isotope analyses reveal that chemolithoautotrophic Gamma- and Epsilonproteobacteria feed a microbial food web in a pelagic redoxcline of the central Baltic Sea. Environ Microbiol 11, 326-337.
Stock, A., Jürgens, K., Bunge, J., Stoeck, T. (2009): Protistan diversity in the suboxic and anoxic waters of the Gotland Deep (Baltic Sea) as revealed by 18S rRNA clone libraries. Aquat. Microb. Ecol.: 55: 267-284.
Grote, J., Jost, G., Labrenz, M., Herndl, G. & Jürgens, K. (2008). Significant chemoautotrophic activity of Epsilonproteobacteria in marine pelagic redoxclines as determined by MICRO-CARD-FISH. Appl Environ Microbiol,74 7546-7551.
Jost, G., Zubkov, M. V., Yakushev, E., Labrenz, M. & Jürgens, K. (2008). High abundance and dark CO2 fixation of chemolithoautotrophic bacteria in anoxic waters of the Baltic Sea. Limnol Oceanogr 53, 14-22.
Grote, J., Labrenz, M., Pfeiffer, B., Jost, G. & Jürgens, K. (2007). Quantitative distribution of Epsilonproteobacteria and a specific Sulfurimonas subgroup in pelagic redoxclines of the central Baltic Sea. Appl Environ Microbiol 73, 7155-7161.
Hannig, M., Lavik, G., Kuypers, M.M.M., Woebken, D., Martens-Habbena, W. & Jürgens, K. (2007). Shift from denitrification to anammox after inflow events in the central Baltic Sea. Limnol. Oceanogr. 52: 1336-1345
Falk, S., Hannig, M., Braker, G., Wardenga, R., Köster, M., Jürgens, K., Gliesche, C. (2007). NirS containing denitrifier communities in the water column and sediment of the Baltic Sea. Biogeosciences 4: 255-268
Labrenz,M., Jost, G. & Jürgens, K. (2007). Distribution of abundant prokaryotic organisms in the water column of the central Baltic Sea with an oxic-anoxic interface. Aquatic Microbial Ecol 46, 177-190.
Hannig, M., Braker, G., Dippner, J.W. & Jürgens, K.(2006.). Linking denitrifier community structure and prevalent biogeochemical parameters in the pelagial of the central Baltic Proper (Baltic Sea). FEMS Microb. Ecol. 57: 260-271.
Labrenz,M., Jost, G., Pohl, C., Beckmann, S., Martens-Habbena, W., & Jürgens, K. (2005). Impact of varying in vitro electron donator/-acceptor conditions on potential chemolithoautotrophic communities from marine pelagic redoxclines. Appl Environ Microbiol 71, 6664-6672.
Diploma/Master/Bachelor theses
Andreas Rogge (2012): Investigations on stable isotope labeled substrate enrichments in “Sulfurimonas gotlandica” strain GD1 using the NanoSIMS technology. Masterarbeit, Universität Rostock.
Marlen Marcuse (2012): Ökophysiologische Charakterisierung von Choanoflagellaten-Isolaten aus der Redoxkline der zentralen Ostsee. Masterarbeit, Universität Rostock.
Juliane Wendt (2012): Physiological investigations on the carbon metabolism of a chemoautotrophic epsilonproteobacterium (“Sulfurimonas gotlandica” str. GD1). Masterarbeit, Universität Rostock.
Ramona Kirsch (2011): Saisonalität der Expression prokaryontischer amoA-Gene in pelagischen Redoxklinen der zentralen Ostsee. Diplomarbeit, Universität Rostock.
Carlo Berg (2010): Identification of active microorganisms in pelagic redoxclines of the central Baltic Sea. Diplomarbeit, Universität Rostock.
Katrin Kießlich (2008): Diversität verschiedener CO2-Fixierungswege in einer pelagischen Redoxkline der zentralen Ostsee. Diplomarbeit, Universität Rostock.
Carsten Wunsch (2008): Quantifizierung von Genen und Transkripten des reversen TCA-Zyklus im Tiefenprofil pelagialer Redoxklinen der zentralen Ostsee. Diplomarbeit, Universität Rostock.
Kathrin Mundt (2008): Quantifizierung und Identifizierung von heterotrophen und autotrophen Mikroorganismen in der pelagischen Redoxkline des Schwarzen Meeres. Diplomarbeit, Universität Rostock.
Falko Toetzke (2007): Depth distribution, composition and concentration of archaea within a pelagic redoxcline. Bachelor thesis.
Sabrina Beckmann (2006): Anaerobe chemolithoautotrophe Diversität, Quantität und Aktivität in einer pelagischen Redoxkline. Diplomarbeit, Universität Rostock.
PhD theses
Ruth Anderson (2012): From single species to the community level: exploring under-researched avenues in protist ecology and protist-prokaryote interactions. PhD thesis, Universität Rostock.
Sabine Glaubitz (2010): Distribution and activity of chemolithoautotrophic microorganisms in Baltic pelagic redoxclines. PhD thesis, Universität Rostock.
Jana Grote (2009): Aut- and synecological characterization of a potential chemolithoautotrophic key organism of the central Baltic Sea. PhD thesis, Universität Rostock.
Michael Hannig (2006): Structure and function of denitrifying bacteria in the water column of the central Baltic Sea. PhD thesis, Universität Rostock.