
Diversity, Distribution and Ecology of Aquatic Protists
In contrast to bacteria and archaea (prokaryotes), protists are single celled organisms that have real nuclei (eukaryotes), in most cases mitochondria or mitochondrial derivates and sometimes also chloroplasts. They form a paraphyletic group of contemporary microorganisms with the development of multicellularity in several evolutionary lineages therein (slime molds, plants, animals, fungi). Protists are ubiquitous distributed and constitute much of the genetic diversity within the eukaryotes despite their generally diminutive size. Their extreme diversity encompass an incredible range of sizes, forms, and functions with pivotal roles in aquatic microbial food webs and ecosystem functioning. For example, photosynthetic protists are major contributors to the biomass and primary production in euphotic water masses. Heterotrophic nanoflagellates (“HNF”) and smaller ciliates participate in controlling and shaping the diversity and activity of prokaryotic communities via grazing.
We try to answer some of the open questions concerning protist diversity, ecology and protist-bacteria interactions using different methods and approaches. We focus mainly on the Baltic Sea but participate for comparison also in studies on other marine and limnic systems.
Comprehensive investigations on protist diversity, particularly on nanoflagellates, need similar molecular techniques as for prokaryotes and have until now only sporadically been performed for the Baltic Sea. Therefore no information is currently available whether similar phylogenetic groups of protists as recently found globally in the ocean are present in the Baltic Sea. Besides classical microscopical techniques, we apply fingerprinting techniques (DGGE, SSCP), clone libraries and fluorescent-in-situ hybridization (FISH) to analyse the diversity and composition of aquatic protist communities. The recently developed massively parallel 454 pyrosequencing technique provides the opportunity to rapidly obtain large quantities of sequences at low costs and without cloning bias. For unicellular eukaryotes, several primer sets were tested on two variable regions of 18S rRNA so far: V9 (typically about 150-200 bp) and V4 regions (approximately 400-500 bp). With each of these markers a wide range of taxonomic groups had been detected.
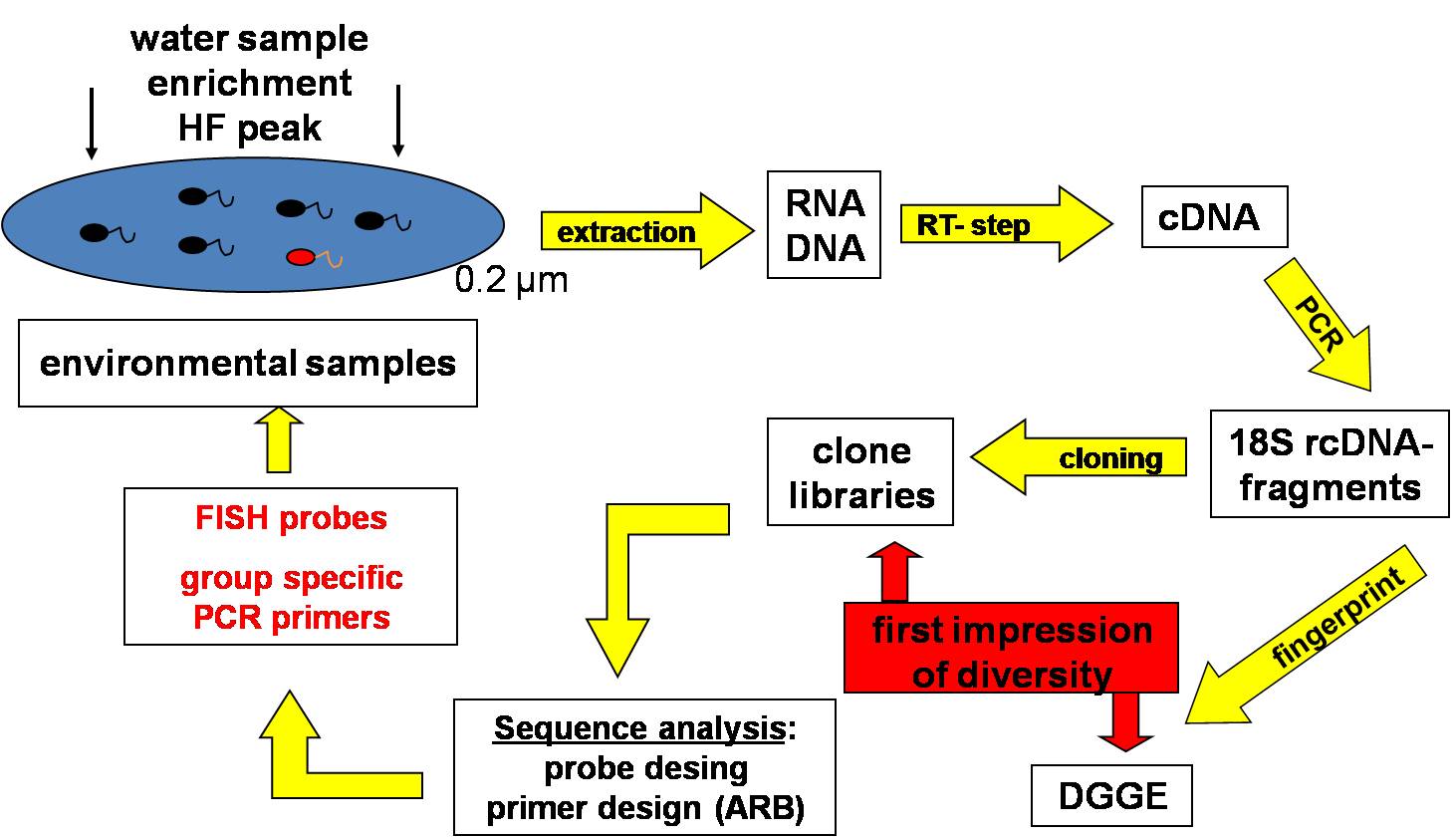
Diversity, phylogeny and ecological role of heterotrophic protists in the redox gradients of the central Baltic Sea
Our knowledge regarding the diversity and function of microorganisms in pelagic redoxclines is based mainly on data from prokaryotic communities. Comparable studies on microbial eukaryotes (protists) are still largely lacking. Our aim is to investigate the composition and functional role of phagotrophic protists (mainly flagellates and ciliates) along the redox gradients. To this aim, the quantitative species composition of phagotrophic protists is being examined by a combination of molecular techniques (18S DNA/RNA fingerprints, clone libraries, in situ hybridization, quantitative PCR), classical microscopical methods, and incubation experiments with natural assemblages and isolated organisms. In order to understand the impact of the oxygen gradients on the major mortality factors for planktonic prokaryotes, we also quantify mortality rates due to protist grazing and viral lysis along the gradients. The Baltic Sea redoxcline will serve as a model system to reveal a general understanding of bacteria-protist interactions at oxic-anoxic interfaces. Samples and experiments from other marine redoxclines will serve to test whether the patterns observed in the Baltic Sea can be generalized.
In situ investigations on redoxcline protist communities were are successfully complemented with studies on isolated organisms from the environment, which are used for ecophysiological investigations and species descriptions of morphological indistinguishable species. For example, we were able to isolate and cultivate several choanoflagellate strains belonging to lineages so far only known from environmental sequencing approaches. Quantitative experimental data for choanoflagellates from Gotland and Landsort Deep (Baltic Sea) indicate their preference for oxygen-depleted zones and thus their potentially high ecological relevance for Baltic Sea hypoxic waters. Cultivation of protists inhabiting hypoxic habitats has been unsuccessful in most cases, and has never yet succeeded for choanoflagellates, even though these small bacterivorous flagellates are known to be ecologically relevant in terms of biomass and abundance. Our preliminary ecological investigations have shown that one of the choanoflagellate strains is able to grow successfully under suboxic conditions in the laboratory and reach high cell abundances by very weak oxygen saturation.
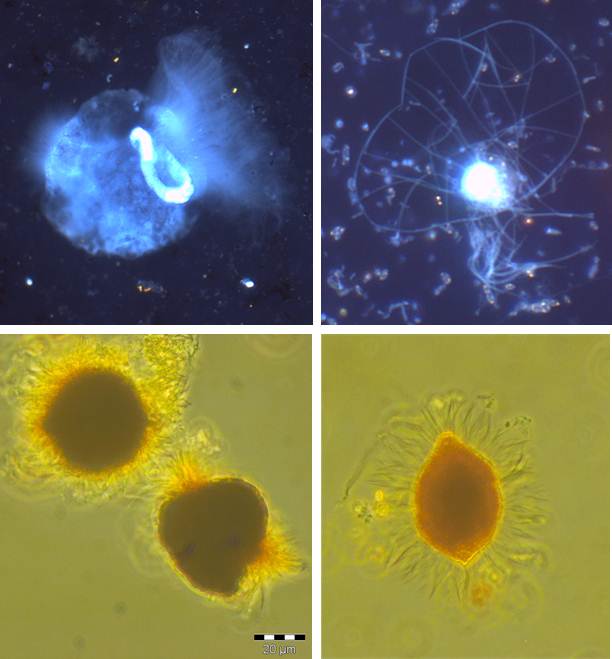
Diversity and ecology of benthic protozoa along environmental gradients (salinity, oxygen)
Based on their global distribution and abundance, and their ability to adapt to a broad spectrum of abiotic factors, protozoa play a major role in food webs and biogeochemical pathways in aquatic as well as terrestrial systems. For the pelagic zone the interaction of aquatic protists (especially heterotrophic nanoflagellates and ciliates) with different trophic levels and the implications for carbon- and nutrient cycles are well described (“microbial loop”). It is very likely that within benthic systems protists play a similar essential role. However, the knowledge of the effect of multiple environmental factors on their diversity and abundance as well as the functional meaning of the different groups of protists is still scarce.
Due to its different gradients the Baltic Sea is predestined as sampling site to address these topics. On the one hand, the salinity gradients (vertical and horizontal) allow us to examine community compositions from marine over brackish to freshwater sites, on the other hand, different levels of oxygen over the whole sampling site give us the chance to investigate the impact of oxygen availability on benthic protistan communities.
The aim of this research is to identify benthic key species according to changing abiotic factors to further investigate the linkage between changes in community structure and biogeochemical processes, closing the present gap between pelagic and benthic communities. For this, samples of the surface sediment were taken along a salinity gradient across the whole Baltic Sea, and along oxygen gradients through the deep basins. The samples are classified according to the sediment structure (particles size and sorting), CNS ratios and chemical composition of the pore water. The analysis of the microbial eukaryotic diversity follows the 18S approach. After Extraction of the RNA, samples are amplified via PCR and subsequently analysed by 454 pyrosequencing and phylogenetic sequence alignment. Quantitative analysis of cellular abundance is assessed by fluorescence microscopy.
IOW Press Release of 9 August 2012
The smallest predators in the Baltic Sea
Bacteria-consuming unicellar organisms, the heterotrophic flagellates, are known to be the smallest predators in the oceans. To shed light onto the vastly unresolved biodiversity of the Baltic Sea's tiniest hunters – researchers of the IOW did just the opposite: they turned off the light!
A better understanding of ecosystem functioning requires a close examination of the complex interactions between organisms that constitute the food web of a given ecosystem. But this leads to a fundamental question: Who eats whom? Who is the predator, who is the prey? Under water, the classical interactions between predator and prey play out on a miniature scale: between unicellular microorganisms. Unquestionably, the most successful predators acting at the very base of the marine trophic pyramid are the so-called heterotrophic flagellates – tiny unicellular organisms that are propelled by whip-like organelles (flagella) and which prey on even smaller bacteria.
Predacious flagellates have long (since the 1980s) been known to massively impact bacterial growth, through grazing pressure, and to be a relevant link between bacterial production and higher multicellular organisms such as zooplankton or fish. Yet, many questions remain to be answered, including: Who are the key players in the hunt for bacteria? Which species comprise the natural assemblage of heterotrophic flagellates?
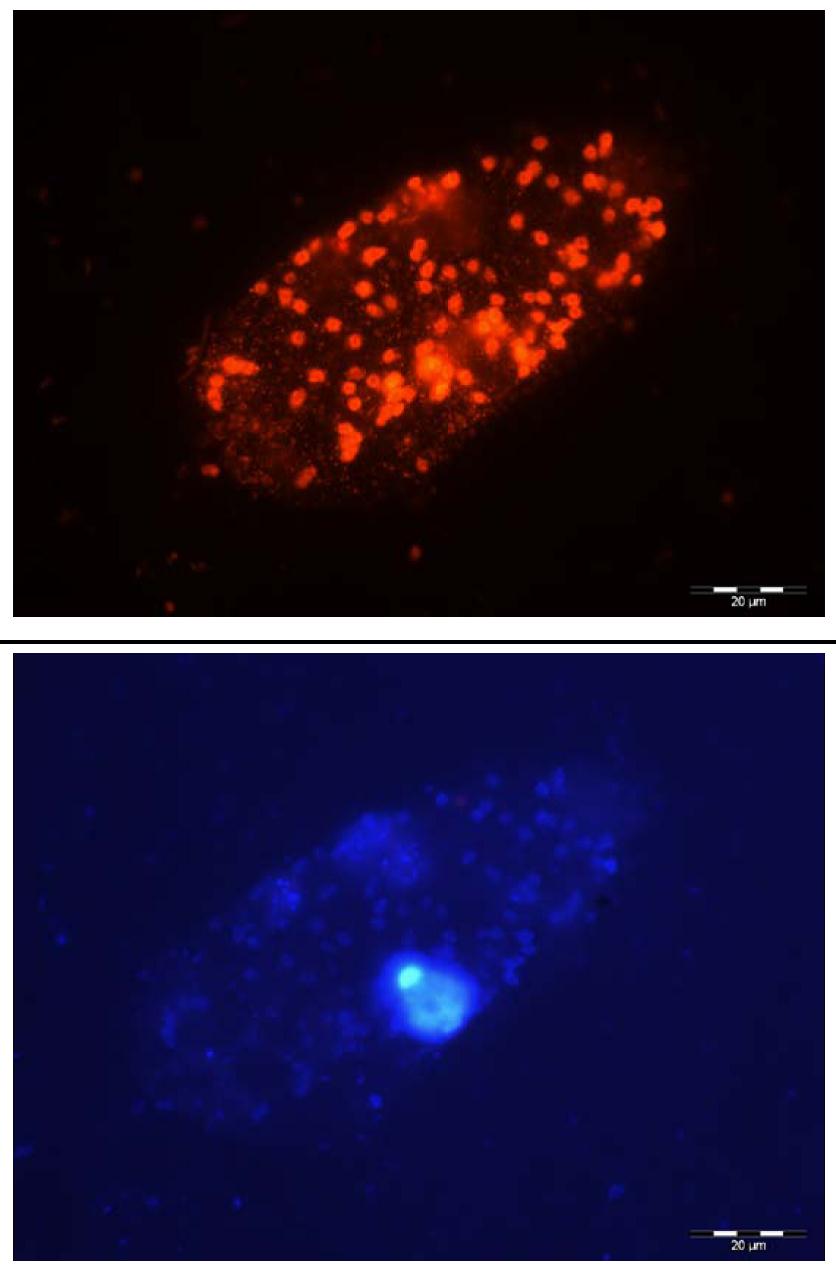
Marine scientists associated with the team of Prof. Dr. Klaus Jürgens, from the Leibniz Institute for Baltic Sea Research Warnemünde (IOW), and his colleagues from the Institut de Ciències del Mar (ICM) in Barcelona are the first to successfully obtain deep insight into the largely unknown species diversity of predatory flagellates in the Baltic Sea, identifying a range of novel and undescribed species. "Flagellates belong to the domain of protists and as they are only between 2 and 20 μm in size single species are hardly distinguishable under the microscope", says Felix Weber, the first author of the study. "For that reason, molecular methods have been applied for over a decade to investigate natural protist assemblages and to describe the species inventory of various aquatic systems."
However, such methods, which are based on the characterization of certain genes (18S ribosomal RNA), lead to the detection of bacterivorous protists as well as their photosynthetically active counterparts. "This implies a fundamental drawback of commonly used molecular methods since a protist detected in the water sample exhibits its ‘genetic identity’ whereas the functional role played by the protist in the ecosystem at the time of sampling remains concealed, i.e., whether it is a predator or synthesizes its biomass through the power of sunlight, just as green land plants do", explains Felix Weber.
To avoid this problem, the team came up with a simple solution: The water sample was left to stand for a while in the dark. The underlying trick was that these conditions acted as a "functional filter" on the protistan community: light-dependent phototrophic flagellates declined in number whereas bacteria-consuming species increased in abundance. "After one week of incubation, the ratio of phototrophic to heterotrophic organisms reached a value favourable for the characterization of the tiny predators on the basis of their 18S rRNA genes," states Weber. "Especially by applying a molecular comparison of the community composition at the start and end of the incubation, we were able to draw conclusions about which taxa developed in the dark and thus are very likely bacterial consumers."
Thus, the scientists identified various as-yet-undescribed flagellate taxa and simultaneously addressed their ecological role. "In the Baltic Sea, prey – in this case meaning bacteria - have already been subject to intense studies," says Klaus Jürgens. "But this most recent investigation was the first to gain important insights into the biodiversity of the smallest predators in the Baltic Sea – the heterotrophic flagellates."
Furthermore, according to the results of the IOW and ICM scientists, the community composition of the small predators in the brackish southern Baltic Sea very closely resembles a fully marine system such as the North Sea. The crucial question, whether the predator composition changes within the steep salinity gradient of the Baltic Sea, similar to the observed shifts of other organisms in the Baltic, will be the subject of further research.
The results were published in the article:
Weber F., del Campo J., Wylezich C., Massana R. & Jürgens K. (2012) Unveiling Trophic Functions of Uncultured Protist Taxa by Incubation Experiments in the Brackish Baltic Sea. PloS ONE, DOI:10.1371/journal-pone.0041970.
This study was partly funded by the German Academic Exchange Service (DAAD), (F2100GKSD), http://www.daad.de, and the German Science Foundation (DFG) (JU 367/11-1).
Contact:
Felix Weber, Biological Oceanography, IOW
(Tel.: 0381 / 5197 252, Email: felix.weber@io-warnemuende.de)
Dr. Barbara Hentzsch, Public Relation, IOW
(Tel.: 0381 / 5197 102, Email: barbara.hentzsch@io-warnemuende.de)
Nils Ehrenberg, Public Relation, IOW
(Tel.: 0381 / 5197 106, Email: nils.ehrenberg@io-warnemuende.de)
Publications
2012
● Weber, F., Campo, J.d., Wylezich, C., Massana, R., Jürgens, K. (2012): Unveiling trophic functions of uncultured protist taxa by incubation experiments in the brackish Baltic Sea. PLOS One 7: e41970. doi: 10.1371/journal.pone.0041970
● Dunthorn, M., Klier, J., Bunge, J. and Stoeck, T. (2012), Comparing the hyper-variable V4 and V9 regions of the small subunit rDNA for assessment of ciliate environmental diversity. J. Euk. Microbiol. 59: 185–187. doi: 10.1111/j.1550-7408.2011.00602.x
● Engel M, Behnke A, Klier J, Buschbaum C, Volkenborn N, Stoeck TEffects of the bioturbating lugworm Arenicola marina on the structure of benthic protistan communities. Mar. Ecol. Prog. Ser. (in press)