
Impact of Salinity on Microbial Communities
As physical determinant, salinity has a biological relevance by influencing media density, which again requires osmoregulatory functions of organisms. Already Remane (1934) postulated that brackish settings could be species-poor and typicallyinhabited by organisms near the limits of their tolerances. He described an asymmetric ideal curve distribution for benthic invertebrates in the Baltic Sea with a minimum in species diversity between a salinity of 5–8. Analogously, zooplankton composition of the Baltic Sea (with exception of the Gulf of Bothnia) seems to have also a clear “Remane-relationship” (Schiewer, 2008). Also for phytoplankton, the Darss Sill (salinity approximately 9) has been defined as an important biological borderline (Kell, 1973; Wasmund et al., 2004; Witkowski et al., 2005) with marine species dominating westwards and freshwater species dominating eastwards.
Since the first saltwater-freshwater bacterial community comparisons have been conducted (Nold and Zwart, 1998; Glöckner et al., 1999), comparable patterns in the shift of major bacterial groups have been shown in various estuaries at different geographic locations, e.g., Delaware Bay (Cottrell and Kirchman, 2003) and Chesapeake Bay (Bouvier and del Giorgio, 2002) in the USA, or Pearl River in China (Zhang et al., 2006). All studies recognized drastic shifts in bacterial community composition along the estuaries, from the prevalence of Betaproteobacteria in freshwater regions to dominance of Alphaproteobacteria in the higher salinity regions. Actinobacteria seem to follow the same trend as Betaproteobacteria and decrease with increasing salinity (Kirchman et al.,2005) whereas the abundance of the Cytophaga-Flavobacterium (CF) cluster do not show a clear relationship with salinity.
References
Microbial Diversities in the Baltic Sea Salinity Gradient
Although studies in river outlets could deduce some general patterns in the development of prokaryotic composition during transition from fresh- to saltwater, fast salinity changes that can occur at the scale of hours and together with other factors (e.g. turbidity, temperature changes, nutrient changes) prevent the development of an autochthonous bacterial community. The Baltic Sea, in contrast to most estuaries, is a more stable environment that provides an ideal system to investigate autochthonous brackish bacterial communities. The Baltic Sea is characterized by its distinct horizontal and vertical pelagic salinity gradient, with the unique feature that 62 % of its surface zone is represented by brackish conditions (5-8 ‰ salinity). Saline inflows produce a lateral surface salinity gradient throughout the whole Baltic Sea with high salinities (about 25) in the transitions zone of the Kattegat and low salinities (around 3) in the Gulf of Bothnia. With an estimated residence time of more than 20 years (Reissmann et al. 2009), the Baltic Sea is an optimal habitat to investigate the distribution and function of microbial brackish communities and to examine shifts in composition and functions along salinity gradients.
Single observationsconcerning the diversity of pelagic microorganisms in the Baltic demonstrated the presence of marine as well as freshwater taxa, but currently no coherent view exists on the area of a shift between marine and freshwater microorganisms, or the composition of potential brackish water microbial assemblages. It is also unclear if, analogously to higher organisms, the microbial diversity in brackish water is significantly reduced in comparison to marine or freshwater systems. It is our hypothesis that the Baltic salinity gradient modifies the microbial composition and also affects distinct microbial functions in the ecosystem. To verify this hypothesis, we sampled transects covering different seasons and the horizontal and vertical salinity gradient of the whole Baltic. Bacterial diversity has been examined applying 16S rRNA/rRNA gene Tag pyrosequencing analyses, resulting in the first biogeographical as well as microbial richness mapping of the Baltic Sea on a high resolution, three-dimensional basis. In a first study, we assessed bacterial community succession along the salinity gradient of the Baltic Sea, using 454 pyrosequencing of partial (400 bp) 16S rRNA genes of 213 samples collected along vertical profiles at 60 sampling stations during a summer situation.
Principal coordinates analysis based on β-diversity separated the samples largely according to salinity indicating that salinity is the major determinant for the bacterial composition for our samples. Along the investigated salinity gradient a change in the bacterial composition was manifested at broad phylogenetic levels (Fig. 1) as well as at fine-scale phylogenetic levels (Fig. 2), with closely related populations occupying different salinity and depth ranges. A major shift in the bacterial communities in the surface water was observed at salinity 8–10 and at salinity 3–4. Between these abiotic barriers, the bacterial community was composed of a diverse combination of freshwater and marine groups, along with populations unique to the brackish environment. Because of the hydrological characteristics of the Baltic Sea, the bacterial community identified in the central Baltic Sea cannot be the result of simple freshwater and saltwater mixing, but reveals the first detailed description of an autochthonous brackish microbiome.
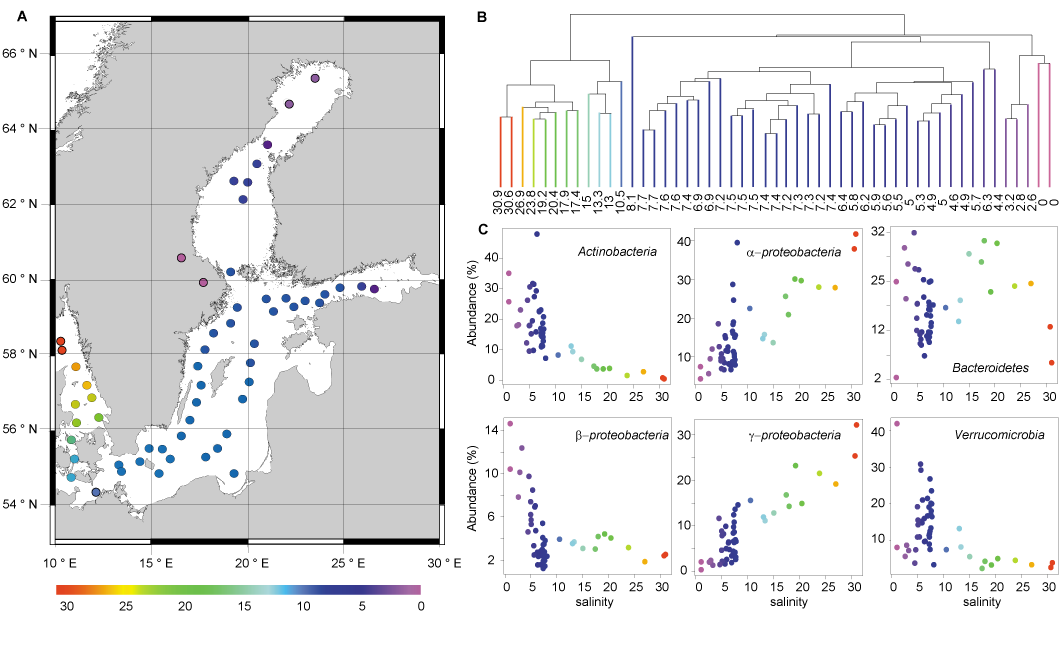
Figure 1: Bacterial communities of surface samples along the Baltic Sea salinity gradient. (A) Map of the Baltic Sea, where each dot represents a sampling station colour-coded according to salinity. (B) Hierarchical clustering of the surface water samples based on bacterial community composition similarity, colour coded according to salinity. The samples cluster in three salinity ranges: freshwater 0–3.2 (purple), brackish 4.6–7.7 (blue), and marine-brackish 10.5–30.9 (other colours). The samples having a salinity of 6.3 and 4.4 were sampled outside Stockholm and St. Petersburg, respectively, the two largest cities in the region, and may potentially be influenced by anthropogenic emissions of nutrients and/or pollutants. (C) Relative abundance vs. salinity for abundant phyla and proteobacterial classes colour coded according to salinity. Modified from Herlemann et al. (2011).
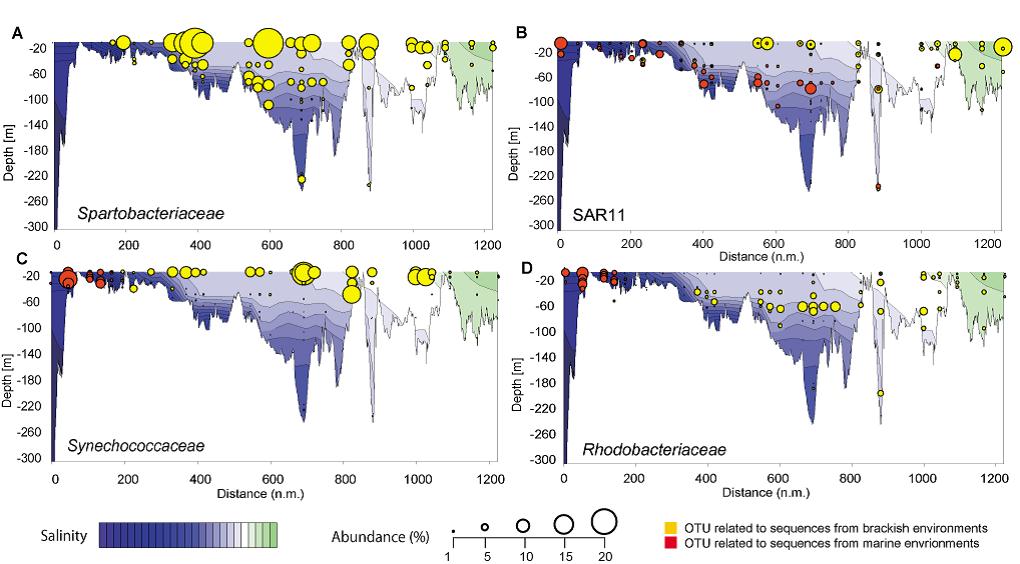
Figure 2: Spatial distribution and abundance of operational taxonomic units (OTU) along a Baltic Sea transect extending east of Gotland and into the Bay of Bothnia. (A) Two dimensional plot of the most abundant OTU in our study, which is related to Spartobacteriaceae. Populational shifts from marine to freshwater populations along the salinity gradient of the Baltic Sea of SAR11 (B), Synechococcaceae (C), Rhodobacteriaceae (D). Modified from Herlemann et al. (2011).
The investigation of the overall diversity along the salinity gradient revealed that analogous to the situation for marine animals, the number of obtained sequences matching to a marine 16S rRNA gene database increased with increasing salinity, while the number of sequences matching to freshwater sequences decreased (Fig. 3). Similar to the Remane curve, and in accordance with the clustering results above, the major shift between freshwater and marine biomes occurred around salinity 8, which suggests that marine bacteria tolerate a larger salinity spectrum (8–32) than freshwater bacteria (0–8). In the Central Baltic Sea (salinity 6–10), a roughly equal proportion of sequences matched both environments, which suggests that the community has originated to equal extents from the two biomes. In contrast to benthic faunal diversity, a lower bacterial diversity was not observed at intermediate (brackish) salinity levels, which suggests that the rapid adaptation of bacteria has enabled a diversity of lineages to fill what for higher organisms remains a challenging and relatively unoccupied ecological niche.
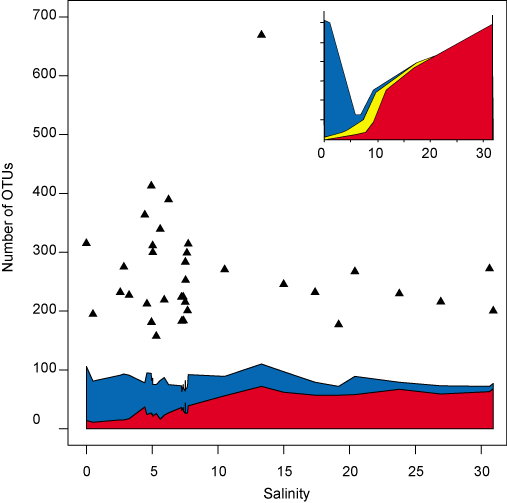
Figure 3. Comparison of the sequences of operational taxonomic units (OTUs) from surface water in our study to environmental sequences from freshwater and marine reference environments outside of the Baltic Sea. Blue and red areas indicate the Number of OTUs ≥ 97% identical to freshwater (blue) and marine (red) sequences, respectively. Black triangles are estimated number of OTUs based on the CHAO1 richness estimator. Inset: The Remane curve (reconstructed from Remane, 1934) showing benthic fauna richness along a Baltic Sea salinity gradient. Blue, number of freshwater species; red, number of marine species; and yellow, number of brackish specialist species.
Publications resulting from our research on salinity gradients
Herlemann, D.P.R., Labrenz, M., Jürgens, K., Bertilsson, S., Waniek, J.J., Andersson, A.F. (2011): Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 5: 1571-1579.
Diploma/Master/Bachelor theses
Benjamin Bergen (2011): Quantification of Baltic Sea bacteria by catalyzed reporter deposition-fluorescence in situ hybridization. Diplomarbeit, Universität Rostock.
Jana Woelk (2011): Distribution of potential photoheterotrophic bacteria in the salinity gradient of the Baltic Sea. Diplomarbeit, Universität Rostock.